REAPR: Realignment for Prediction of Structural Non-coding RNA
The REAPR pipeline performs computational screens for structural
non-coding RNA while revealing even strongly structurally misaligned
RNAs. It is the first such pipeline that is sufficiently fast to
screen entire eukaryotic genomes.
The REAPR pipeline is freely available and licensed under GPL 3.0. We provide online installation instructions (including download links of required software) and documentation.
The REAPR pipeline and its application to fly and the human ENCODE region is described in
Please find supplementary information here.
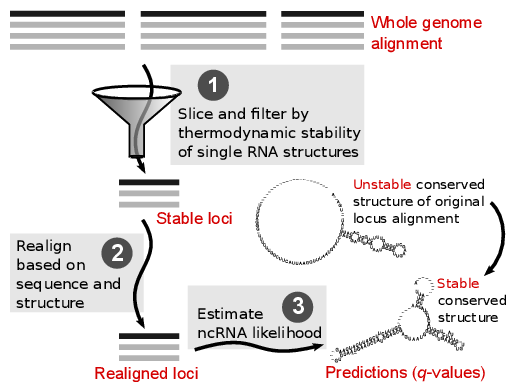
The REAPR pipeline consists of three steps: (1) The screened
whole genome alignment is sliced into windows. The windows are
filtered based on an alignment independent criterion for structural
RNA: thermodynamic stability of single RNA structures. The stable
windows are merged into stable loci. (2) Each stable locus is
realigned based on sequence and structure similarity. This results in
a stable locus alignment that correctly aligns RNA structure even when
the locus was originally misaligned. (3) A conventional ncRNA
predictor is applied to estimate ncRNA likelihood from the corrected
locus alignment. Whereas a locus of true ncRNA shows only unstable
conserved structure in the original alignment, if this structure was
misaligned in the whole genome alignment, the pipeline can reveal the
stable conserved structure of the ncRNA.