BetaWrapPro
The BetaWrapPro web server is now operational. Please direct
any comments or questions about the program to betawrap@csail.mit.edu.
Submit your sequences to the
BetaWrapPro server.
About BetaWrapPro
BetaWrapPro is a program for prediction and comparative modeling of the
beta-helix and beta-trefoil protein motifs. It accomplishes this by profile
wrapping.
Please see the help page for more information.
If you use informtion from this website, or results from the BetaWrapPro program, please cite:
A.V. McDonnell, M. Menke, N. Palmer, J. King, L. Cowen, B. Berger, "Prediction and comparative modeling of sequences directing beta-sheet proteins by profile wrapping". Proteins: Structure, Function, and Bioinformatics 2006, 63: 976-985.
High-scoring sequences
Here is a list of the best-scoring beta-helices detected by BetaWrapPro from SwissProt.
Here is a similar list for the beta-trefoils.
Authors
Links to our home pages:
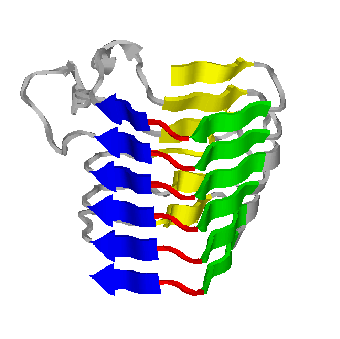
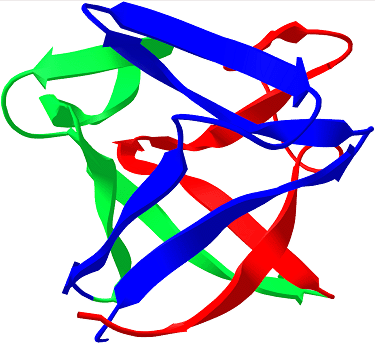
The beta-helix shown is a Rasmol image of Pectate Lyase C from Erwinia chrysanthemi, whose structure was solved by Yoder, Keen, and Jurnak in 1993 (PDB ID 1pcl). The beta-trefoil is a DeepView image of Human Basic Fibroblast Growth Factor (PDB ID 1bff), solved by Kastrup, Ericksson, Dalboge, and Flodgaard in 1993.
Not to scale.


