Supplementary Information for:
R. Singh, J. Xu, and B. Berger. "Struct2Net: Integrating Structure into Protein-Protein Interaction Prediction." Proc 11th Pacific Symposium on Biocomputing 11: (2006): 403-414.
- Predictions on Less-Characterized Proteins:
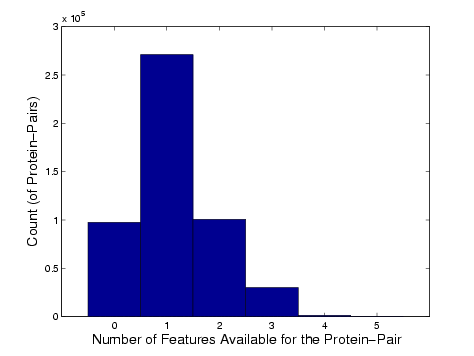
A list of the 1000 less-characterized proteins is here. These proteins were chosen because relatively little data is available for them. As mentioned in the paper, our method uses 6 existing genomic/proteomic features: co-expression, co-essentiality, co-location, similarity in GO terms, similarity in MIPS terms, and interacting domains. For the set of less-characterized proteins, we counted the number of features with available data for each possible protein-pair. The histogram of the number of available features (for each protein-pair) is shown below. About 94% of the pairs have 2 or less features available, i.e. had atleast 66% of their feature set is missing. All the protein-pairs had atleast 2 missing features.
Clearly, use of structure-based methods would be valuable here. Using only the structure-based method, predictions were made for all possible protein-pairs in this set and the top 2000 scoring pairs (as per logistic regression) were chosen. This set of pairs is here.
Why top 2000? (and not, say, top 200): Based on current estimates, we assume that the yeast interactome has about 30,000 interactions. Also, yeast has about 6000 proteins. Suppose the size of the yeast interaction graph scales linearly with the number of proteins; then the size of a 1000-protein sub-network will be 30K * (1000/6000) = 5000. If the size of the interaction graph scales quadratically with the number of protein then the size of a 1000-protein sub-network will be 30K * (1000^2/6000^2) = 833. The true yeast interaction network is a scale-free network that scales somewhere between linearly and quadratically, with number of nodes. Thus, the number of interactions in a 1000-protein sub-network should be somewhere between 833 and 5000. We chose a cutoff between the two: 2000.
- Disease-Related Proteins:
A list of yeast homologs of human disease related proteins was retrieved from here. We searched for interactions involving these in the set of 1000 proteins mentioned previously. Brief discussion for two of the proteins is in the paper. Here, we describe results for other genes:
Yeast Gene Disease related to the Human homolog Brief Disease Description Predicted Interactions Comments PAT1 Adrenoleukodystrophy (ALD) ABC transporter; neurodegenerative disease 26 Set of predicted interactors enriched for lipid and fatty acid transport RAD28 Cockayne syndrome, (CSA) Transcription-coupled repair;progressive neurological dysfunction;photosensitivity 19 Many DNA repair proteins in the set of predicted interactors PEX7 Rhizomelic chondrodysplasia punctata Peroxisomal biogenesis disorder 25 YAT1 Carnitine palmitoyltransferase Lipid metabolism defect; cardiomyopathy 19 Set of predicted interactors enriched for protein-misfolding related proteins and chaperones TPI1 Triosephosphate isomerase Chronic hemolytic anemia and neuromuscular disorders 16 Set of predicted interactors enriched for hexose and monosaccharide metabolism ADE13 Adenylosuccinate lyase Purine nucleotide biosynthesis defect; autism features 4 - Predictions on Entire Yeast Genome: We used the structure-based method, without any other functional information, to perform an all-vs-all prediction of interactions over the yeast genome. Predictions were made involving 6213 genes. Logistic regression was used to rank the predicted interactions. The set of experimentally-determined interactions between members of this proteins is here (gzipped file); it has 13062 predictions. The number of true interactions is likely to be larger. However, for comparison purposes, we chose only the top 13000 predicted interactions. This set is here (gzipped file).
| Questions or comments? Please contact struct2net@csail.mit.edu |