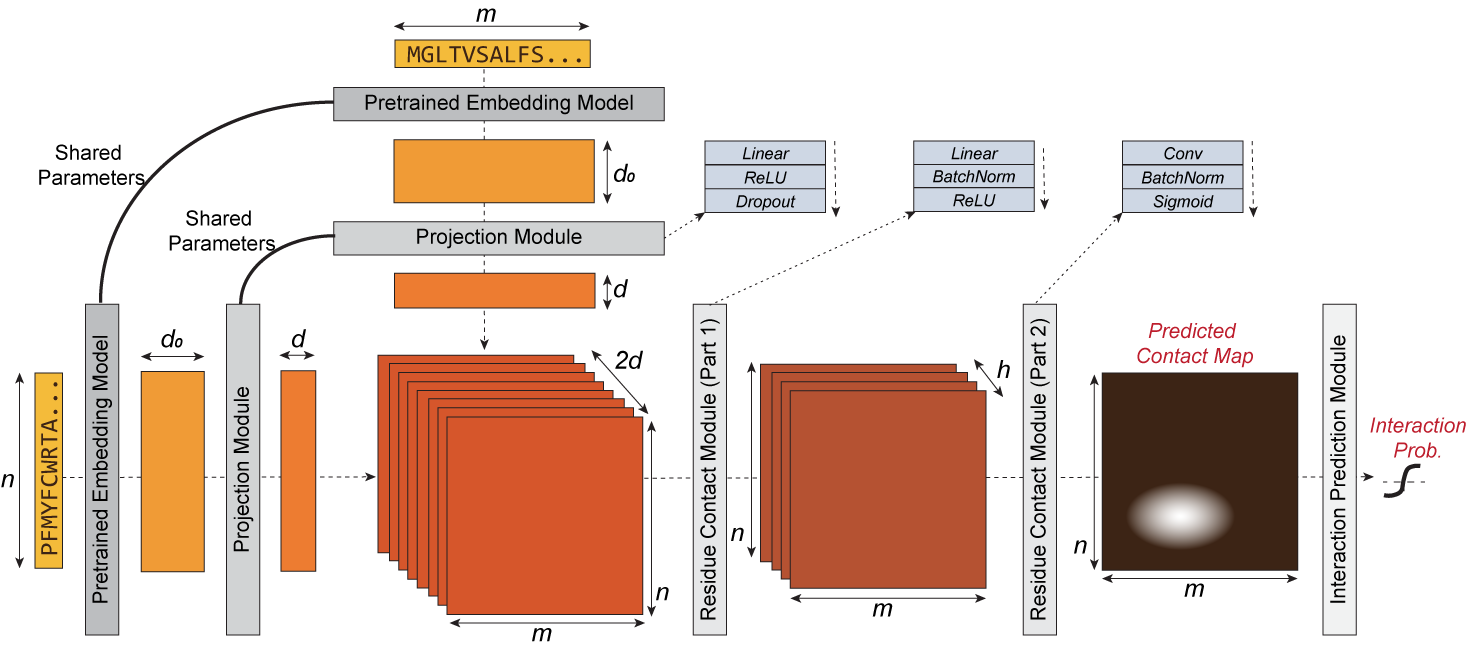
D-SCRIPT is a deep learning method for predicting a physical interaction between two proteins given just their sequences. It generalizes well to new species and is robust to limitations in training data size. Its design reflects the intuition that for two proteins to physically interact, a subset of amino acids from each protein should be in contact with the other. The intermediate stages of D-SCRIPT directly implement this intuition, with the penultimate stage in D-SCRIPT being a rough estimate of the inter-protein contact map of the protein dimer. This structurally-motivated design enhances the interpretability of the results and, since structure is more conserved evolutionarily than sequence, improves generalizability across species.
D-SCRIPT is described in the paper “D-SCRIPT translates genome to phenome with sequence-based, structure-aware, genome-scale predictions of protein-protein interactions” by Sam Sledzieski, Rohit Singh, Lenore Cowen and Bonnie Berger.