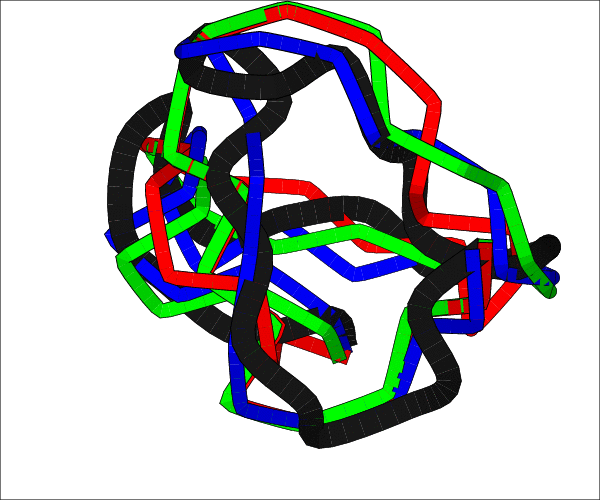
 ChainTweak
ChainTweakChainTweak is a program for sampling the neighbourhood of a given protein backbone. It poses the problem of finding conformations which have the same bond lengths, bond angles but are within a few Angstrom (say 4A) of the original conformation. It does this by using sliding window approach to perform local modifications to the structure without modifying the distal ends.
ChainTweak is described in a paper presented at the Pacific Symposium of Biocomputing at the Big Island, Hawaii (Jan 2005):
Singh R. and Berger B. ChainTweak: Sampling from the Neighbourhood of a Protein Conformation. Proceedings of the 10th Pacific Symposium on BioComputing, 2005.
Supplementary Information for this paper is available here.
Linux binaries for the program are here. Both the interface and the internals *will* change as this program is still being actively worked on-- it's part of our research! We are working on making it more user-friendly and are looking into making the source code available.
******************** NEW! ************************************** Modeling Variability in Crystallographic Structures Xtal. **************************************************************
updated : 24-Jul-2009 15:19